Leading up to the UseR! 2016 conference, I had been looking forward to attending Yihui Xie’s tutorial on R Markdown - there were a few things I had wanted to ask him on how to make custom formats using html_document().
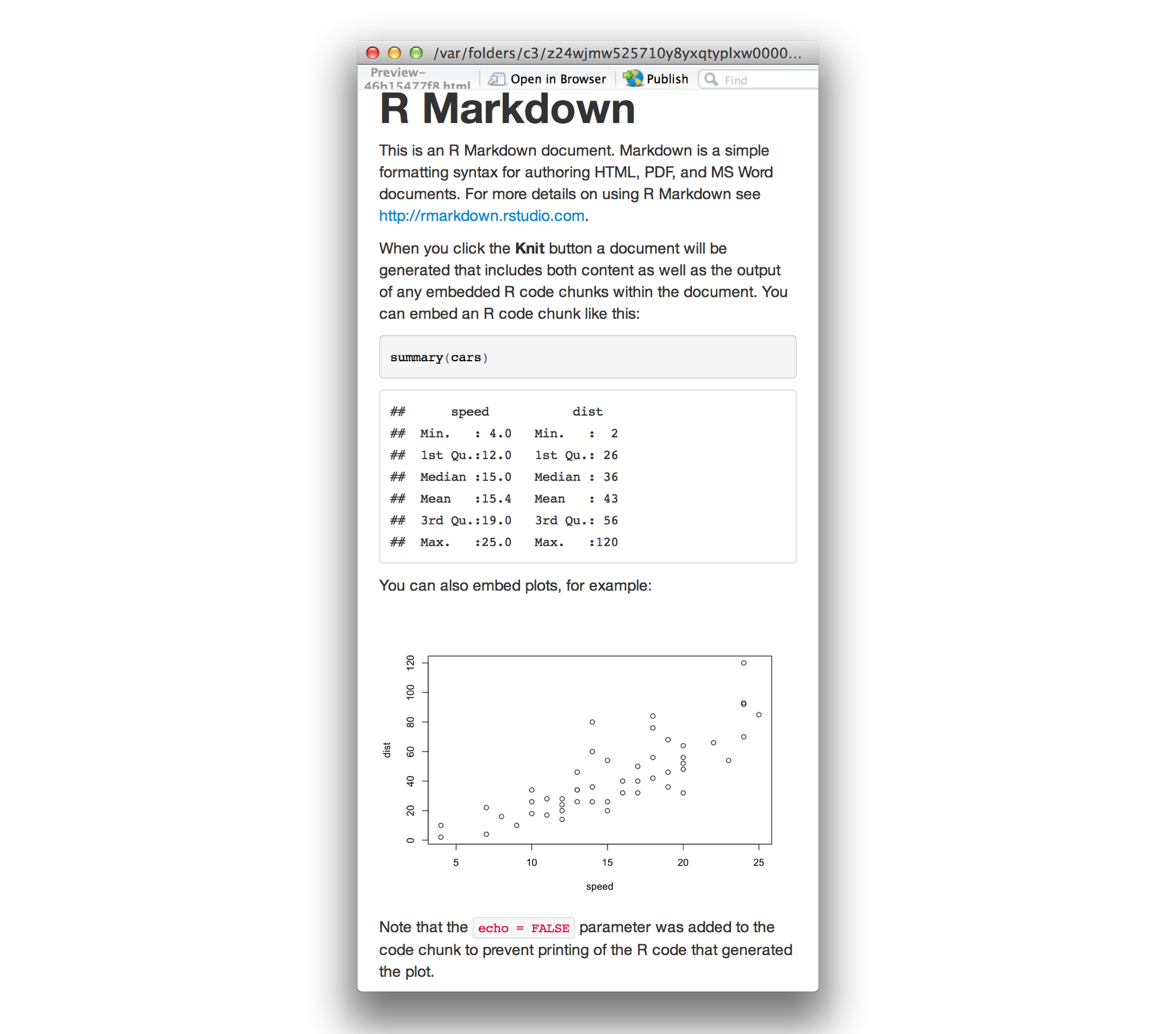
- Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents. For more details on using R Markdown see. When you click the.Knit. button a document will be generated that includes both content as well as the output of any embedded R code chunks within the document.
- One of these packages is the markdown-preview, which gives Atom the ability to save markdown content to HTML. Best of all, markdown-preview is already installed by default! The first step to convert Markdown to HTML is to open your Markdown document in Atom. Then toggle the Markdown preview by pressing the CTRL+SHIFT+M keyboard shortcut.
Join Charlie Joey Hadley for an in-depth discussion in this video Publishing R Markdown HTML to the web, part of Creating Reports and Presentations with R Markdown and RStudio.

Then, a funny thing happened. A few days before the conference started, I got an email from Yihui, sent also to Karl Broman. Yihui’s visa had been delayed, so he could not attend the conference: could one or both of us deliver his tutorial material?
Given everything Yihui has done for the community, the only possible answer was “yes”. So Karl and I divvied up his slides and each up us did our own deep-dive into our material. As it happened, I was to deliver the material that dealt with my questions - so this was a first-rate opportunity to get it figured out!
In days before the tutorial, with help from Yihui, I got most of it worked out, and in the days and weeks to follow, we sorted out the rest. This page is a distillation of what I had been wanting to figure out.
As I write the rest of this out, I remember fondly the four (somewhat panicked) days of getting everything sorted out with Karl and Yihui. A word of thanks here to Karl for putting together the framework that let us bring tutorial in for a landing. Also, thanks to Yihui for thinking of me as someome not completely unsuitable to fill in. And a huge thanks to the participants in the tutorial for their patience.
References
RStudio has a great series of articles:
Yihui’s book.
Get the tutorial slides, etc.
This is a great place to get started on the concepts I try to demonstrate here.

Further: https://github.com/juba/rmdformats
Packages
First, it important to note that this is based on the dev version of R Markdown - pending the next CRAN release.
To follow along here, you will need to install a package, user2016docdemo, that has a series of functions that return custom formats.
Examining the format
It will be a lot easier to figure out how customize things if we understand (a little bit) what is going on. Even though R Markdown is capable of so much more, for the purposes of this discussion we will assume that you are interested only in producing an html file.
Format list
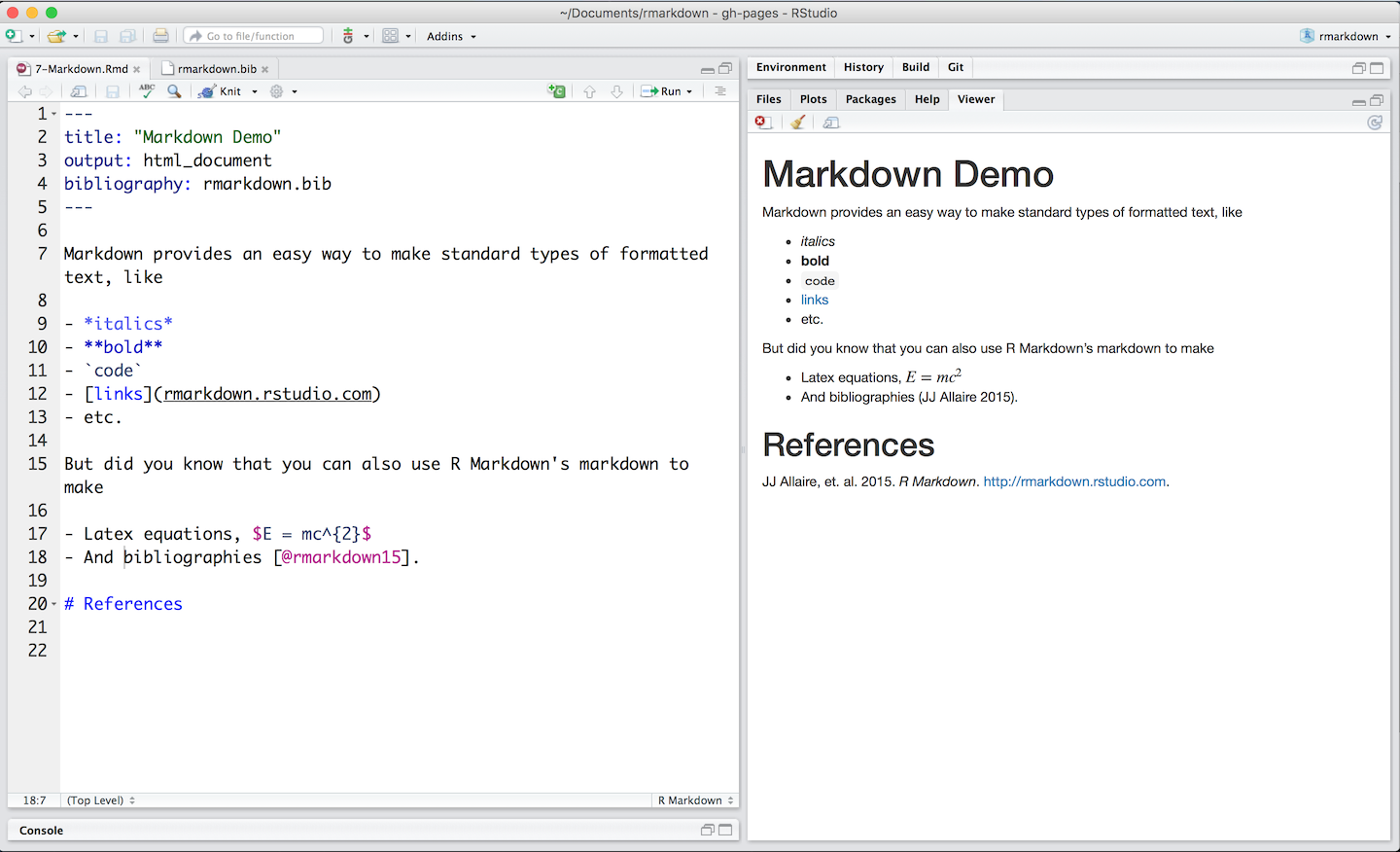
The rmarkdown::render() function has two jobs: (1) to knit the R Markdown file into a Markdown file, (2) to use pandoc to turn the Markdown file into an html file. In order to do this, render() needs a set of instructions - this set of instructions is a format, and it is stored as a list.
The purpose of the rmarkdown::html_document() function is to help you to generate this list. Using Kent Russell’s listviewer package, let’s look at the default format returned by html_document() function.
For our purposes, we are most-interested in the first parts of the list, entitled “knitr” and “pandoc”. You may wish to click around to see what’s what.
Format function
R Markdown Html To Word
The job of any R Markdown format function, including your custom format functions, is to return the format list.
These should look familiar, as they are these are parameters that you use to put in a yaml header at the start of an R Markdown document. For example:
When rmarkdown::render() is run (either from the knit button, or from the command line), it passes along the yaml information “under” html_document: to the rmarkdown::html_document() function. It bears repeating that the best way to figure out how to construct your yaml header is to look at the help page for html_document().
As you design your custom formats, keep in mind that you will be able to pass arguments to your format function using the yaml header.
Custom formats
The biggest biggest mental-block (as least for me) to making an easily deployable custom-format is building a package. The reason this is necessary is that when we hit the knit button in RStudio, the render() function runs in a new R session - this is for a very good reason. The only way I know to get the render() function the “list of instructions” is to provide a package function that returns it.
Package
Luckily, Hadley has made it a whole lot easier for all of us to develop packages. I’d highly recommend his book on building packages, available online: http://r-pkgs.had.co.nz/.
Here’s a minimal list of steps.
- open an empty project
devtools::create('.')devtools::use_package('rmarkdown')devtools::use_readme_rmd()devtools::use_mit_license()(if that’s your thing)use_github()(assumes you have a github.com account)- write and save some custom functions (rest of this section)
- hit the build button to install locally
- commit and push to GitHub every so often
Simplest possible (do nothing)
The simplest possible custom format is a a function that simply passes everything on to rmarkdown::html_document(). Here’s the source code for the simplest-possible custom-format function, one that simply passes everything along to rmarkdown::html_document()
When we look at the list of instructions returned by the default, we see that it is (hopefully) no different than before.
At some point, I’d like to put some screenshots here of a sample page - or maybe just link to the results.
Change some defaults
One simple thing that you can do is to change some of the defaults of the html_document() format:
Similary, we can look at what the function returns:
It is not immedately apparent that there is a difference between the output of html_doc_0() and html_doc_1(). Being both stubborn and not-so-bright, I spent way too much time tracking down where we can find the difference. Here it is: the format list has some elements that are functions; functions have environments. Therein lies the difference. Let’s look at the environment of one of the functions returned as a part of html_doc_1():
We see our information in the args element, code_folding has a value of 'show' and theme has a value of 'readable'.
It is probably more useful to demonstrate what this does, rather than show the details of environments, so put a link here eventually.
Convert Markdown Html
Make your own theme

Here’s where we can have some fun. If the standard themes are not what you need (or want), you can create your own bootstrap theme at http://bootstrap-live-customizer.com/.
This is a re
Session info
Encountering errors attempting to knit your RMarkdown documents on CISER servers? Check out the answers below to solve.
Knit HTML & Word
Error message received when I attempt to knit RMarkdown file to HTML or Word.
pandoc.exe: : openBinaryFile: does not exist (No such file or directory)
Error: pandoc document conversion failed with error 1
Execution halted
Example 1
R Markdown Html To Pdf
#GREEN TEXT IS A COMMENT. CREATED FOR YOUR REFERENCE
#SOLUTION: Copy and paste the normal colored font commands included below. Make sure to check that output from your commands resemble mine.
#Check out library paths. I want to C: drive path to be listed first.
library(rmarkdown)
.libPaths()
#[1] 'rschfs1x/userRS/F-J/jrg363_rs/Documents/R/win-library/3.6'
#[2] 'C:/Program Files/R/R-3.6.1/library'
#Switch order of paths using commands below.
myPaths <- .libPaths() # get the paths
myPaths <- c(myPaths[2], myPaths[1]) # switch them
.libPaths(myPaths) # reassign them
#View result of .libpaths(), results should be switched.
.libPaths()
#[1] “C:/Program Files/R/R-3.6.1/library”
#[2] “rschfs1x/userRS/F-J/jrg363_rs/Documents/R/win-library/3.6”
Example 2
#Check out library paths. I want to C: drive path to be listed first.
.libPaths()
#[1] 'rschfs1x/userts/jrg363_ts/Documents/.checkpoint/2019-12-14/lib/x86_64-w64-mingw32/4.0.1'
#[2] 'rschfs1x/userts/jrg363_ts/Documents/.checkpoint/R-4.0.1'
#[3] 'C:/PROGRA~1/R/R-40~1.1/library'
mypaths <- .libPaths()
#Take 3rd and 2nd paths in that order. ignore 1st.
mypaths1 <- c(mypaths[3], mypaths[2])
#Set Lib paths
.libPaths(mypaths1)
.libPaths()
Now knit file to HTML or Word should work. If not please email *protected email* with questions.
Information gotten from following URL
Rstudio libraries
Knit PDF
Error message received when I attempt to knit to pdf.
Sorry, but C:PROGRA~1MIKTEX~1.9miktexbinx64pdflatex.exe did not succeed.
! The log file hopefully contains the information to get MiKTeX going again:
! C:Usersjrg363_rsAppDataLocalMiKTeX2.9miktexlogpdflatex.log
Error: LaTeX failed to compile program.tex. See https://yihui.org/tinytex/r/#debugging for debugging tips. See program.log for more info.
Execution halted
#This process of setting up PDF took me an hour to load. You must do this process every time you open a new Rmarkdown session and would like to knit to pdf. Suggested idea to setup your RMarkdown to knit to pdf, then disconnect your session. Thus signing in to that same server will have you all set up. View URL below for how to disconnect.
#Manage CISER Sessions
#SOLUTION: Copy and paste the normal colored font commands included below.
install.packages('tinytex')
#Users can not write to C: drive. If pop-up windows asks question included in screenshot below answer 'YES'
#Command below takes the most time. Approx an hour to run to completion. Once this command is complete running user is allowed to knit to pdf.
tinytex::install_tinytex()
#While the above command runs expect to see the following windows pop-up. Errors suggest noting, they can be ignored. Answer 'OK' for all these windows.
#Output of command used above too long to paste.
#Use command below to test if above command worked. Expect result to be TRUE
tinytex:::is_tinytex()
#[1] TRUE
You should be able to knit to pdf now.
Any issues or questions please send email to *protected email*
Information received from following URL
knit to pdf
